Evolution
Evolution
Having a BLAST: The Torley Saga, Cont.

Editor’s note: Updated on 2/21/18.
There has been an ongoing discussion between Dr. Vincent Torley, a philosopher writing at Uncommon Descent, and myself on the subject of pseudogenes, synteny, and common descent. These are technical terms and the arguments may seem abstruse, but the topic is important because of its implications.
Those who hold to common ancestry accept that, at the very least, each major group of animals is descended from a common ancestor, and some hold that the common ancestry goes all the way back to the first cell. The question at hand then is whether we share common ancestry with chickens and opossums, and by implication, with chimpanzees. What we have been discussing through all these posts is this: How strong is the evidence that we have the remnants of an egg yolk protein, vitellogenin, in our genome? Do we come from an egg-laying ancestor?
The precipitating event for the entire discussion was a post, “Vitellogenin and Common Ancestry: Reading Tomkins,” by Professor Dennis Venema of Trinity Western University, over at BioLogos. Venema claimed evidence for an egg yolk pseudogene in humans, based on a paper “Loss of Egg Yolk Genes in Mammals and the Origin of Lactation and Placentation” by David Brawand, Walter Wahli, and Henrik Kaessmann. My apologies if you are just now entering the conversation. I posted a critique of Venema’s post, “The Vitellogenin Pseudogene Story: Unequally Yolked.” Then Vincent Torley weighed in, as coached by Professor Josh Swamidass of Washington University, on why my analysis was wrong, wrong-headed, intellectually intransigent, and unreasonable. Torley’s post was called “Consider the Opossum.”
The theme of that post was the evidence for common descent and its predictive nature — in contrast to common design, which according to Torley is ad hoc as an explanation and not predictive. The argument in Torley’s post went something like this: “We know we are related by common descent, so there must have been an ancestor where placentas began to form and egg yolk usage began to decline. Therefore we would expect to find egg yolk proteins left over in the genomes of mammals. And we did.”
In my first post, “The Vitellogenin Pseudogene Story: Unequally Yolked,” I outlined my reasons for thinking that the evidence for a vitellogenin pseudogene was weak in humans. This argument was based on the available evidence from the original paper by Brawand et al., mentioned above, that reported the original alignments of the syntenic vitellogenin regions of chick, opossum, and human. (Synteny refers to the alignment of DNA, typically between species, in the same order and with identifiable sequence identity.) That paper reported that there was a vitellogenin gene remnant in human DNA in the region where it should be. Unfortunately they published very little data as the basis for their claim, and I didn’t think it substantiated that claim.
In response, in his post “Consider the Opossum” Torley quoted some alignment numbers for human and chicken sequences from a computer scientist named Glenn Williamson. These alignments did not appear to match the Brawand et al. data as reported. In fact, based on what was reported, I couldn’t tell what they looked like. In my response, “Vincent Torley Thinks I Have Egg on My Face,” I asked where Williamson had gotten his numbers, since I could find no evidence of such an alignment in the paper.
Williamson replied and reported his alignment online. Since then, all four parties concerned, Torley, Swamidass, Williamson, and I, have exchanged information about the vitellogenin (VGT as it is now abbreviated) alignment with human DNA. Williamson’s alignment, which was the source of the numbers Torley reported, was as he said. Swamidass did his own set of BLAST (Basic Local Alignment Search Tool) alignments and sent them to me, and I did one BLAST alignment myself. All of us agree that the VGT alignments are more extensive than was reported in Brawand et al.
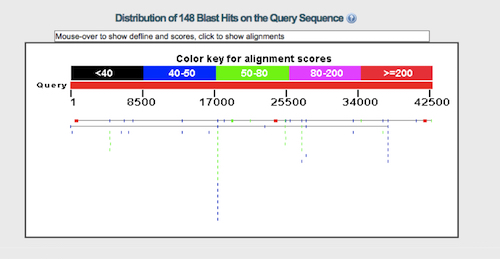
The number of alignments varies depending on precisely how the BLAST program is run, but by my alignment there are three areas of roughly 75 percent identity, each between 360 and 483 bases in length. The amount of sequence aligned is between 3 and 5 percent of the total gene’s length, counting exons and introns. To illustrate the degree of alignment I have reproduced the graphic image from my BLAST alignment below. (Click on it for a closer view. It matches the alignments done by Josh Swamidass pretty well.) The thin horizontal line represents the chicken VTG gene. The colored lines are regions where the human sequence aligns with the VTG gene. The red spots are the areas of good alignment, while the blue and green spots are generally poorer alignment.
The distribution of hits is similar to what Brawand et al. reported in their dot plot, given that they were probably using more sensitive alignment criteria than I was. They are also similar to alignments Williamson and Swamidass showed me. The size of the hits are similar, though Swamidass and Williamson find a few more hits than I do. Since the alignment is so weak, we would need to run the BLAST program many times to get a better idea of which alignments are best. Brawand et al. ran two thousand alignments and reported the best one.
There is definitely more similarity between chicken and humans than the Brawand paper reported. Why they didn’t report the full degree of alignment I don’t know.
Still, despite the increased alignment, this is on the borderline of what is detectable as a match. The fact that things vary from alignment to alignment indicates the match is weak. If there ever was a human vitellogenin gene, there’s almost nothing left of the original gene. Swamidass tells me that given the long time that has passed since our last common ancestor with chickens, this is to be expected. Yes, I know. But there is an alternate explanation — the possibility that a vitellogenin gene was never there to begin with — though I know Swamidass and Torley will vigorously disagree with me.
I wrote my original post to critique Venema’s claim of strong evidence for a human vitellogenin pseudogene. That critique still stands. His illustration greatly overestimates the degree of similarity.
But enough of vetting this human VTG pseudogene. We’ll have to agree to disagree. In my next post I will continue the discussion about vitellogenin genes and Torley’s critique of my argument, but this time focusing on opossums, whose alignment provides much stronger evidence for a VTG pseudogene.
Clarification
The alignments mentioned above, when considered by themselves, are statistically significant. Though not definitive, they are consistent with the remnants of a vitellogenin gene, which would argue for common descent. The alternative explanation of common function is still possible. Distinguishing between the two hypotheses might be possible by first examining all mammalian genomes for similar syntenic, aligned sequences. Deletion of such aligned sequences in mouse, for example, would have no effect if the alignment is due to common descent, but may have an effect if the sequence is functional.
Photo credit: © Vidady — stock.adobe.com.