 Evolution
Evolution
 Intelligent Design
Intelligent Design
The Deeper Issues of the Worm Video Debate: Ursula Goodenough’s Hypothesis
In response to the Discovery Institute video “How to Build a Worm,” distinguished evolutionary biologist Ursula Goodenough at Washington University has proposed a solution to the puzzle we raise. Her solution fails, but its failure is instructive, because it reinforces the main point of the video: Whatever brought the process of animal development into existence must have been a cause with foresight.
I met Ursula Goodenough in February 2006, when she, Michael Ruse, and I participated in a symposium on evolution and design at Missouri Western State University. Although she thought design was misguided at best, her biological knowledge and intellectual candor were very appealing, so I was happy to see her weigh in on the worm video, and to float a hypothesis.
1. Close but no cigar.
Here’s what she had to say at Jerry Coyne’s Why Evolution Is True, divided into its major explanatory components. (Goodenough was writing a blog comment, not a formal article, so there’s a certain looseness to her hypothesis, which required me to supply details that she did not. If she has a tighter formulation, of course, I’d love to see it. I should also note Ann Gauger has pointed out to me that how one explains the origins of multicellularity will shift, depending on the eukaryotic starting point one chooses, and that’s true — but I’m using Goodenough’s own starting point, at least as far as I can discern it.)
Background assumptions:
The common ancestor to the 3 major radiations — bacteria, archaea, and eukaryote — was already a pretty sophisticated dude, notably with the ability to switch gene expression on and off in response to internal and external cues. Hence the ancient single-celled organism that engaged in the first multicellular experiment leading to animals already had a full “tool-kit” of genes/proteins/regulatory elements and a sophisticated capacity to deploy their expression differentially in TIME. The multicellular trick, which evolved independently at least 20 times in different radiations (e.g. land plants do it very differently than animals), is to ALSO deploy differential gene expression in SPACE, that is, to construct an organism with more than one cell and then express a given gene subset in one cell type and a different subset in another. In modern animals this is triggered by patterning the egg such that some of the cells that arise during early embryonic cleavage wind up with portions of the egg cytoplasm that send them in one direction and others to go in a different direction, much as Nelson describes in the movie.
- Goodenough’s model step #1: Begin with a unicellular protist that can switch between ordinary cell reproduction (mitotic division) and gamete or sexual reproduction (meiotic reduction).
So how can we model the origin of the first multicellular creature in the animal lineage circa 600 million years ago? Let’s take a unicellular species that switches back and forth in time between being a dividing cell and a gamete, a switch triggered by an environmental variable like nitrogen availability. Many modern eukaryotic protists fit this description.
Goodenough didn’t name which protists she had in mind, but the choanoflagellate species Salpingoeca rosetta (for instance) has been reported to undergo an environmental switch in ploidy (haploid to diploid) and exhibits a sexual life cycle (Levin and King 2013). Given that choanoflagellates are thought to be the unicellular sister group to the animals (Fairclough et al. 2013), this seems a reasonable candidate. The target for the origin of metazoan multicellularity will be a creature with diploid (somatic) cells and haploid (gamete or sex) cells.
- Goodenough’s model step #2: The daughter cells remain attached, with only one cell capable of meiotic reduction / gamete formation.
What happens next is that instead of two daughter cells splitting off from one another, they stay together — not very hard to model — and one cell is capable of switching on the sex genes and the other is not — again not hard to model. The mutations generating these outcomes are random, but they occur in the context of regulated genomes that are highly responsive to mutational tweaking. It’s not like coming up with something from scratch.
- Goodenough’s model step #3: Mutation and selection can now elaborate from this two-celled beginning, increasing cell number and adding features.
The point is that all we need to do is posit that there WAS some adaptive feature of this arrangement and the game is on. Increasingly elaborate proto-animals can now evolve from this template animal via additional mutations, sticking 4 and then 8 cells together, organizing additional cell types, etc., each innovation either enhancing the original adaptation or initiating additional adaptations.
Figure 1 depicts Goodenough’s model:
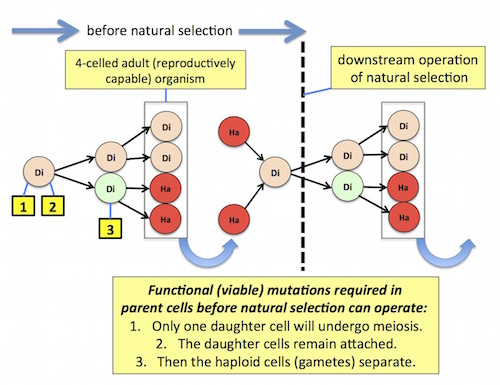
Figure 1. Click on the image for a larger view.
Notice that at least three coordinated mutations are required before natural selection can operate: (1) unequal daughter cells are produced, with only one capable of meiotic reduction / gamete formation, (2) instead of separating, the daughter cells remain attached, and (3) after meiotic reduction occurs, producing haploid gametes, only the gametes separate from the parent organism. (These mutations are listed here strictly by their required functional outcomes; the actual mutations in real, not toy, organisms, might involve many molecular changes, at multiple sites.)
Goodenough didn’t specify gamete separation, but that function would be absolutely necessary for sexual reproduction and successful fertilization (i.e., returning to the diploid starting point in the next generation [blue arrow]). If haploid gametes are not free to separate as single cells, to fuse with other gametes, their role in the life cycle will be lost, and evolution will stall out immediately.
Which raises a tricky functional problem, obvious if one studies Figure 1 carefully. Notice that the mutation “daughter cells remain attached,” (2) in the diagram, must somehow be escaped, or reversed by mutation (3), expressed only in the haploid gamete cells, at the very next cell division. Gametes that remain physically connected to their parent organism cannot play their necessary functional role, namely, starting a new physically discrete organism.
But those gametes must also carry the novel instruction for the whole (mutation 2), “cells remain attached,” absolutely required to construct a multicellular organism. Thus, whatever mutation instructs the cells to stay together must simultaneously allow some cells, the gametes, to separate from the parent organism — or be reversed by another mutation (number 3) expressed only in the gametes.
How are these contradictory instructions reconciled, prior to the operation of natural selection? (See the vertical black dotted line in the figure.) Mutations 1, 2, and 3, and doubtless many others, must occur for sexual reproduction in a multicellular organism to arise de novo via natural selection.
Yet, as noted above, reproductive capability, where the traits of the whole organism are transmitted to offspring, is a necessary condition of natural selection itself (Endler 1986). So the construction of multicellularity must occur prior to the operation of selection. Evolutionary biologist Paul Rainey, who works on the origins of multicellularity, calls this “a paradox of quite major proportions” noting that for unicellular collectives “to become more complex they must be able to participate in the process of evolution by natural selection, but to participate in the process of evolution by natural selection they must be more complex.” (Rainey thinks he has found a solution to the problem, by the way; you can check out his ideas here.)
2. Do “snowflake yeast” provide a real-life example of Goodenough’s hypothesis?
Experiments with baker’s yeast (Saccharomyces cerevisiae) in Michael Travisano’s lab at the University of Minnesota bear directly on Goodenough’s “daughters should stick together” hypothesis. Travisano and colleagues selected for yeast cells where mother-daughter separation was disrupted, generating multi-cell “snowflake” phenotypes that they argue lie on the evolutionary pathway to genuine multicellularity. See Ratliff et al. 2015 (open access) for details.
The experiments are fascinating, but they tell a decidedly mixed story. Ratliff et al. found that the snowflake phenotype was caused by mutations to a key regulatory element, the transcription (DNA-binding) factor ACE2. This gene and its protein product control the expression of several downstream genes involved in the precise mechanics of cell separation (see Table 1 of Ratliff et al. 2015, p. 4). Disrupting ACE2 keeps mothers and daughters stuck together, satisfying the first step in Goodenough’s hypothesis, or any hypothesis where the origins of multicellularity require normally single cells to cluster irreversibly.
And there’s the rub. Yeast typically reproduce by budding and mitotic division, yielding haploid daughters from haploid mothers, but under certain conditions (e.g., starvation), yeast can also produce mating-type haploid spores which unite with other spores to form diploid cells (which then undergo meiosis). To function as mating cells, however, these haploid daughters must be able to separate physically from their mothers.
Although Ratliff et al. don’t report this result, it appears very likely that the ACE2 mutation also destroys this wild-type capacity for physical separation, or sporulation (gamete formation), because too many of the proteins required for the physical separation of mothers and daughters have been downregulated by the ACE2 mutation. In others words, the gain of a “multicellular” snowflake phenotype in yeast may have come at the high cost of losing normal haploid spore formation.
One step forward, one bigger step backwards. The second and third steps of Goodenough’s hypothesis, at least as exemplified in yeast, wouldn’t happen.
3. Telling adaptive stories requires attention to functional details before selection can operate.
In text that I didn’t cite, Ursula Goodenough said that her hypothesis would probably be criticized as what she called a “Just-So Story.” But, she continued, we need simply “posit that there WAS some adaptive feature of this arrangement and the game is on.”
But it’s not the selective advantage that matters, when one tries to explain the origin of animal development. As Goodenough observes, one can always come up with some hypothetical advantage post hoc, and supply that to the formula of natural selection.
Moreover, just-so stories in biology aren’t bad because they’re complicated causal narratives. After all, telling a true story, even if very complicated and appealing to unique events, provides a genuine explanation (e.g., the bridge fell because of causal factors A, B, C, and D, in that order). Instead, just-so stories are bad because they usually don’t work when the individual events they comprise are examined, and the requisite details are rarely forthcoming in any case.
Rather, as Figure 1 shows, the challenge is just getting to the point functionally where natural selection actually operates. That’s an engineering problem, and selection will not help.
It can’t, until at least one reproductively capable organismal prototype is ready for testing. Build that first.
References:
Endler, John. 1986. Natural Selection in the Wild. Princeton: Princeton University Press.
Fairclough, Stephen R., Zehua Chen, Eric Kramer, Qiandong Zeng, Sarah Young, Hugh M Robertson, Emina Begovic, Daniel J Richter, Carsten Russ, M Jody Westbrook, Gerard Manning, B. Franz Lang, Brian Haas, Chad Nusbaum and Nicole King. 2013. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biology 14:R15.
Levin, Tera C. and Nicole King. 2013. Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta. Current Biology 23:2176-80.
Ratliff, William C. et al. 2015. Origins of multicellular evolvability in snowflake yeast. Nature Communications 6:6102.
