 Evolution
Evolution
 Human Origins
Human Origins
 Intelligent Design
Intelligent Design
On Human Origins, a Cautionary Tale with "Surprising" Biological Results

Back in the late 1980s and early 1990s, DNA sequencing was sweeping the scientific scene with its power to answer a number of unsolved problems. Among the pioneers, in 1987 Cann et al. used mitochondrial DNA sequences to show that all women were the descendants of a single woman who lived some time around 200,000 years ago. (Similar work was done later for the Y chromosome.) Some subsequently used the idea to claim that this ancestor was Eve.
Francisco Ayala employed the same technique to challenge their analysis, and to demonstrate that we came from many more than two individuals. To do so, he chose to analyze a highly variable gene, HLA-DRB1, that is part of our immune system.
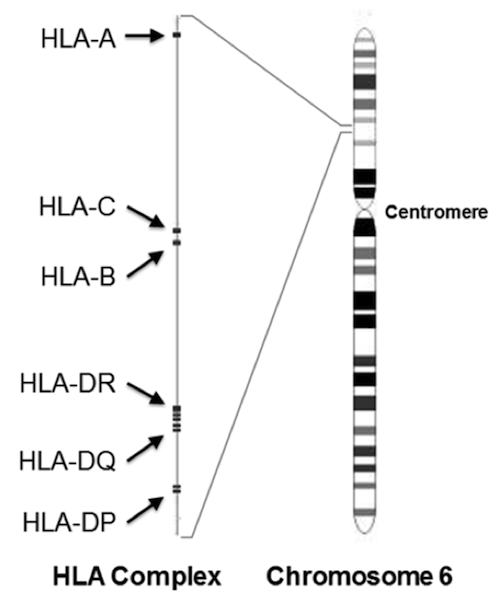
HLA-DRB1 is located in the human leukocyte antigen (HLA) complex. Most of the genes that encode the proteins involved in immune defense are located here. Some of these genes are highly variable in sequence, with hundreds of alleles each.
The variation is mostly limited to one small domain of each protein, called the peptide-binding site (PBS). Each PBS is where the rubber meets the road for recognizing and presenting foreign proteins to the immune system. Variation in the PBS increases the chance that at least some individuals in the population will survive a new parasite or disease; one of these proteins in the population may have a PBS with the right sequence to bind proteins from the attacker. The HLA gene can then direct the immune system to search and destroy anything carrying those foreign proteins. There is also evidence that having different copies of HLA antigens improves fertility.
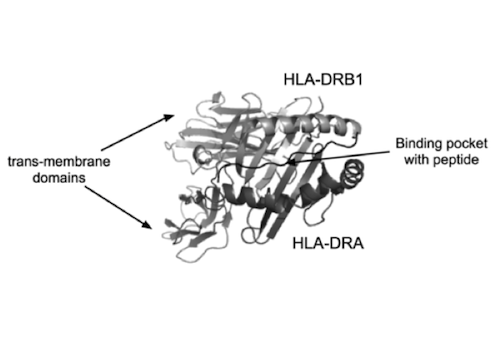
Ayala sequenced the DNA encoding the PBS of HLA-DRB1 precisely because this gene has hundreds of variants in the population. He compared DNA from many Rhesus macaques, chimpanzees, and humans, and based on those sequences, he constructed a phylogenetic tree — a tree intended to show the evolutionary history of this gene.
His conclusion? To account for the current genetic diversity, there had to be at least 32 separate lineages of the gene at the time chimps and humans supposedly diverged from each other. And he explicitly argued that this ruled out Y-chromosome Adam and mitochondrial Eve.
But this wasn’t the end of the story. A few years later a group headed by Tomas Bergstr�m challenged Ayala’s work, saying that he chose the wrong piece of DNA to study. The PBS sequence was inappropriate because it suffered from a high mutation rate and a high rate of gene conversion, both of which will cause an overestimation of the number of lineages for this gene.
Bergstr�m’s group sequenced a neighboring region of the same gene that should not have these problems. They found that the estimated number of HLA-DRB1 lineages dropped from thirty-two to seven.
Now here’s where it gets interesting. The same group later re-sequenced a number of individual HLA-DRB1 genes, but this time they sequenced the whole gene, a monumental task given its size. Guess what they found? Only four lineages predate the supposed split of chimps and humans. A fifth arose about 5 million years ago.
Sequencing the whole gene gave them more complete data, and permitted a finer resolution of the sequence comparisons. It also revealed a number of very interesting things. The introns (the non-coding portions of the gene that get spliced out before it is turned into protein) showed that the four lineages appeared to have been separate for a long time, based on the differences between them. It also showed that there had been an explosion of genetic diversity about 500 to 350 thousand years ago. It was about that time we went from a few lineages of HLA-DRB1 to many, resulting in the incredible diversity we see today.
Let me restate this in case you have missed it. Four lineages of this gene appear to be ancient, because they are so different from one another (there are other possible explanations). But derived from each of those lineages are many, many alleles produced by some process that generates diversity fast. These newly appearing lineages tend to be more similar to one another, indicating that they are young. Surprisingly (there’s that word), their diversity appears to have arisen less than half a million years ago. This result has challenged the strongly held views of some immunologists, and is causing a re-evaluation of the field.
Here are a few take-home lessons:
-
Scientists would be wise not to make strong claims on incomplete data.
-
Science does tend to be self-correcting in the long run.
-
The diversity of the human HLA complex is not due to the ancient preservation of many lineages. Rather, most of the variation is recent.
-
From thirty-two to seven to four. That’s a remarkable journey, one that was entirely unexpected (there’s another of those words). Four alleles can be carried by just two individuals.
I leave you to draw your own conclusions. But don’t be hasty. There’s a whole genome yet to explore.
Image source: Paul Maximus/Flickr.
