 Intelligent Design
Intelligent Design
Replicating DNA with Extraordinary Fidelity: Meet DNA Polymerase
 In a previous article, I gave a brief overview of the complex molecular mechanisms governing DNA replication. Now, I will focus specifically on the replication enzyme DNA polymerase.
In a previous article, I gave a brief overview of the complex molecular mechanisms governing DNA replication. Now, I will focus specifically on the replication enzyme DNA polymerase.
DNA polymerase is the enzyme responsible for synthesizing new strands of DNA, complementary to the sequence of the template strand. The unidirectional DNA polymerase progresses along the template strand in a 3′-5′ direction, since it requires a pre-existing 3′-OH group for the adding of nucleotides. The daughter strand is, consequently, synthesized in a 5′-3′ direction (opposite to the direction of movement of the polymerase since the two strands have an anti-parallel orientation).
There are six different families of DNA polymerases — A, B, C, D, X and Y (Rothwell and Waksman, 2005). These families differ from one another in their design, being specialized for a variety of purposes. For example, DNA polymerase I, found in E. coli, belongs to the A family of polymerases. Beyond its role in finishing DNA replication and removing the RNA primers, DNA polymerase I contains a 5′ to 3′ exonuclease domain, in addition to the 3′ to 5′ exonuclease domain, that allows it to remove nucleotides both in front of and behind it (more on proofreading by exonuclease domains shortly) (Ishino et al., 1995). Polymerases in the B, C and D families are known for their high fidelity (owing to their intrinsic 3′ to 5′ proofreading exonuclease), and are found in eukaryotes, bacteria and archaea respectively. The X family (e.g. eukaryotic polymerases pol ?, Pol ?, Pol ?, Pol ?) plays a role in DNA repair, filling in the gaps created during the process (Yamtich and Sweasy, 2010). Whereas most polymerases cannot replicate past bulky lesions in damaged DNA, the Y family are able to replicate past them (Washington et al., 2010).
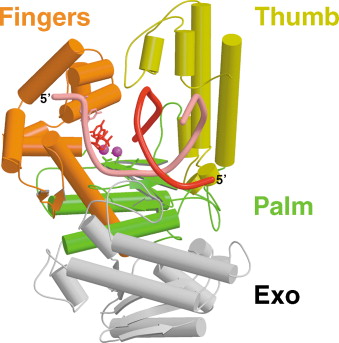
DNA polymerases generally share a common structural framework, with fingers, thumb and palm subdomains that make up the polymerase domain. The diagram below, excerpted from Beard and Wilson (2003), shows the structure of T7 DNA polymerase, revealing its two domains, a polymerase domain (colored) and a proofreading exonuclease domain (gray). The polymerase domain is comprised of three subdomains: fingers, thumb and palm. The finger domain positions incoming nucleoside triphosphates in relation to the template strand. The thumb domain is thought to function in the processivity, positioning and translocation of the DNA, holding in place the elongating DNA duplex. The β-sheet that comprises the palm domain is where the enzyme’s active site lies, which catalyzes the transfer of phosphoryl groups in the phosphoryl transfer reaction.
How does DNA polymerase add new nucleotides to the elongating strand? The polymerase’s active site, found in the β-sheet that makes up the palm subdomain catalyes a phosphoryl transfer reaction. It forms a phosphodiester bond by linking the 3′ hydroxyl group at the end of the template strand to the nucleotide’s 5′ phosphoryl group. The first step in the process is a nucleophilic attack on the 5′-phosphate of the incoming nucleoside triphosphate by the 3′ OH of the growing chain. This reaction releases pyrophosphate (PPi). Within the active site, there are two conserved aspartate residues. The magnesium ions on the carboxylate groups of those aspartates is critical to the reaction. These carboxylate groups co-ordinate the magnesium ions and facilitate their participation in the catalysis by holding them in the right orientation. One of the two magnesium ions activates the 3′ OH group of the terminal nucleotide. The other is responsible for stabilizing a developing negative charge on the leaving oxygen on the incoming nucleoside triphosphate. Side chains on an alpha helix in the finger domain interact with the incoming triphosphate to also stabilize it. Hydrolysis of the pyrophosphate released in this process generates the energy required for driving the reaction forward. For a more detailed review of the mechanisms involved, I refer readers to Rothwell and Waksman (2005).
The process by which DNA polymerase selects the correct nucleotide is less well understood. For a discussion, I refer readers to Markiewicz et al. (2012).
As I stated in my previous post, the rate at which DNA polymerase replicates DNA is thought to be a whopping 749 nucleotides per second (McCarthy et al., 1976) and the error rate for accurate polymerases is believed to be in the range of 10^-7 and 10^-8, based on studies of E. coli and bacteriophage DNA replication (Schaaper, 1993). This extraordinarily high fidelity is accomplished by a remarkable proofreading and error-correcting facility built into the enzyme, which checks the identity of nucleotides both during and after polymerization.
The first level of monitoring occurs by virtue of the fact that, when base-paired with the complementary strand (A with T, or C with G), correct nucleotides precisely fit into the active site, whereas nucleotides that are incorrectly matched will possess a different geometry and will not will not fit so precisely into the active site (Johnson and Beese, 2004).
Sometimes, however, this first level of monitoring will fail to prevent the entry of an incorrect nucleotide. But, thankfully, there is also a second level of proofreading. In addition to the polymerase active site, DNA polymerase possesses a 3′ to 5′ exonuclease active site, which can cleave an incorrect nucleotide from the 3′ end of the growing DNA strand before synthesis of the subsequent nucleotide. When an incorrect nucleotide is mistakenly incorporated, the polymerase’s rate of activity is significantly delayed.
Studies have shown that the presence of mismatch can reduce the polymerase’s efficiency of subsequent elongation by as much as a hundred to a million fold (Kunkel and Bebenek, 2000; Goodman et al., 1993; Echols and Goodman, 1991). This gives enough time for spontaneous denaturing of the DNA at the 3′ end, thereby facilitating the transfer of the 3′ end with the mismatched nucleotide to the polymerase’s 3′ exonuclease site, which catalyzes the removal of multiple nucleotides from the 3′ end of the DNA strand. The 3′ end is subsequently positioned back into the polymerase active site, and the polymerase can then continue its DNA synthetic activity in the 5′ to 3′ direction.
The following animation reveals this remarkable process at work:
It should be noted, however, that not all DNA polymerases possess an intrinsic proofreading exonuclease. Polymerases belonging to the Y family, for example, tend to be significantly less accurate (Friedberg et al., 2001). Members of this family “lack an intrinsic proofreading exonuclease, exhibit low processivity, replicate DNA with low fidelity, and are believed to assist replication complexes stalled at DNA lesions,” (Beard et al., 2002).
DNA polymerase is just one of numerous protein complexes that play an important role in DNA replication. A general overview of the machinery involved in the process of replication is more than adequate grounds to justify a design inference. As we drill down and examine the individual subcomponents that make up the cell’s DNA replication machinery, the case for design becomes ever harder to ignore. In subsequent articles I will continue this exploration of the intricate molecular processes underlying DNA biosynthesis.
