 Intelligent Design
Intelligent Design
 Life Sciences
Life Sciences
DNA Replication: An Engineering Marvel
Recently I have been reviewing some literature on the elegant molecular mechanisms by which DNA is replicated. As an undergraduate biology student, I recall being struck by their sheer complexity, sophistication, and intrinsic beauty. As I read about such a carefully orchestrated process, involving so many specific enzymes and protein complexes, and its extraordinary accuracy, it was almost as though the word “design” jumped off the pages of my textbook and slapped me in the face. The rate of DNA replication has been measured as a whopping 749 nucleotides per second (McCarthy et al., 1976) and the error rate for accurate polymerases is believed to be in the range of 10^-7 and 10^-8, based on studies of E. coli and bacteriophage DNA replication (Schaaper, 1993).
I want to provide here a brief overview of the central processes involved in DNA replication. In subsequent articles, I will examine the individual components in more detail.
DNA Replication is Semi-Conservative
DNA replication is said to be semi-conservative: that is to say, each of the two strands serves as a template for the biosynthesis of a new daughter strand — complementary to the template (Meselson and Stahl, 1958; Parkhomchuk et al., 2009). Thus, two new double helices are produced — each of which possesses one old strand and one new strand.
The Replication Fork
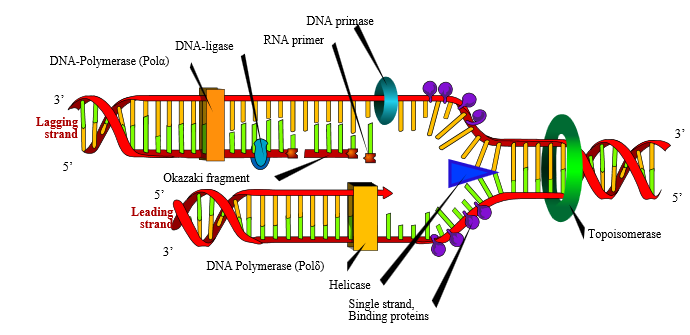
DNA replication commences at particular sites known as origins of replication (ori). Since the replication machinery progresses away from the origins of replication in two directions, a “replication bubble” is formed. At the origins of replication, the DNA double helix is opened and unwound on both sides. This forms what is known as “replication forks.” These replication forks subsequently progress along the DNA in opposite directions, as DNA biosynthesis occurs.
The Initiation Phase
The first step of DNA replication is the initiation phase, a process that is under tight regulation to ensure that it happens no more than one time during cell-division (Mott and Berger, 2007; Nasheuer et al., 2002). This is the stage at which the DNA double helix is opened and unwound to expose the single strands to the enzymes and protein-complexes involved in the replication process. The double-stranded DNA is opened up by an initiator protein. The initiator protein also recruits a specialized class of proteins known as helicases. Helicases are responsible for unwinding the DNA. Once the double-helix has been unwound and opened to form single-strands of DNA, DNA binding proteins associate with the DNA to prevent the single-stranded DNA from winding back on itself.
The Clamp Loader and Sliding Clamp
Two protein complexes, the clamp loader and sliding clamp, are involved in recruiting the DNA polymerase and ensuring that it remains associated with the DNA (Idiani and O’Donnell, 2006; Miyata et al., 2004; Trakselis et al., 2001). The clamp loader is responsible for loading the sliding clamp onto the DNA. After recruiting the DNA polymerase, the sliding clamp literally slides with the DNA polymerase as it moves along the DNA template, keeping them firmly clamped together.
The Elongation Phase
The next stage of the process is known as the elongation phase. This is the stage at which the DNA strands are copied into daughter strands by the replication machinery. The synthesis of the new daughter strand from each of the parent strands is catalyzed by an enzyme complex known as DNA polymerase. DNA polymerase progresses along the template strand, reads the nucleotide bases and adds the complementary nucleotide. The chemistry of this reaction and the polarity of the DNA molecule, however, only permit nucleotides to be added at the 3′ end of the elongating strand. This means that DNA replication can only happen in the 5′ to 3′ direction. DNA polymerase has a remarkably high fidelity, thanks to its built-in proofreading and error-correcting facility.
Synthesis of Primers
The DNA polymerase enzyme is incapable of synthesizing a novel strand from scratch — it is able only to add nucleotides to the 3′ end of already-present nucleotides. Short strands of RNA called primers are therefore synthesized by an enzyme called primase (the RNA primers will ultimately be degraded and replaced with DNA). This raises an interesting design question: Why would a designer engineer DNA polymerase such that it is unable to begin a new strand on its own? RNA polymerase, after all, is perfectly able to perform this operation. What is the design logic in unnecessarily, so it seems, involving the extra steps in synthesizing RNA primers and later removing them and replacing them with DNA? One possible explanation is that the extra stage affords an additional proofreading step — in which case, this apparent extra complexity could be part of a design strategy.
The Leading and Lagging Strands
The two strands of a DNA helix are anti-parallel: that is to say, they have opposite orientations. This raises a difficulty since, as mentioned previously, DNA can only be synthesized in a 5′ to 3′ direction. For one strand known as the leading strand, DNA synthesis is continuous. For the other strand, known as the lagging strand, DNA replication proceeds discontinuously. For replication of the lagging strand to occur, short DNA fragments (known as Okazaki fragments) need to be created constantly from 5′ to 3′, each of them separated by ~10-nucleotide RNA primers. These are then joined by DNA ligase to form a continuous strand.
Relieving DNA Supercoiling
DNA supercoiling is induced by the torsional stress generated by the polymerases and helicases. Strand separation can thus be accompanied by DNA over-winding. A class of enzymes called Topoisomerases relieve this torsional stress by breaking the DNA backbone and relaxing the supercoils. The DNA backbone is then ligated back together. In so doing, this class of enzymes removes the supercoils.
Termination of DNA Replication
DNA replication terminates when the two replication forks, which move in opposite directions, meet. In bacteria such as E. coli, this takes place at particular terminator sequences (ter), which associate with a specific DNA-binding protein called Tus. Terrence A. Brown explains how termination in bacteria occurs:
The mode of action of Tus is quite unusual. When bound to a terminator sequence, a Tus protein allows a replication fork to pass if the fork is moving in one direction, but blocks progress if the fork is moving in the opposite direction around the genome. The directionality is set by the orientation of the Tus protein on the double helix. When approached from one direction, Tus blocks the passage of the DnaB helicase, which is responsible for progression of the replication fork, because the helicase is faced with a ‘wall’ of ?-strands which it is unable to penetrate. But when approaching from the other direction, DnaB is able to disrupt the structure of the Tus protein, probably because of the effect that unwinding of the double helix has on Tus, and so is able to pass by (Figure 13.22B; Kamada et al., 1996).
The orientation of the termination sequences, and hence of the bound Tus proteins, in the E. coli genome is such that both replication forks become trapped within a relatively short region on the opposite side of the genome to the origin (see Figure 13.22A). This ensures that termination always occurs at or near the same position. Exactly what happens when the two replication forks meet is unknown, but the event is followed by disassembly of the replisomes, either spontaneously or in a controlled fashion. The result is two interlinked daughter molecules, which are separated by topoisomerase IV.
Far less is known about termination in eukaryotes, in which there are no specific termination sites. For a discussion of one model of replication-fork termination in eukaryotes, see Fachinetti et al. (2010).
Maintaining Telomeres
Since eukaryotes have linear chromosomes, the DNA polymerase is unable to continue all the way to the end of the chromosomes. This leads to a progressive telomere shortening with each round of cell division. In the germ cells (which pass on their DNA to the next generation), the enzyme telomerase is responsible for maintaining telomere length.
Conclusion
The systems responsible for DNA replication are well beyond the explanatory efficacy of unguided processes involving chance and necessity. Indeed, machinery of the complexity and sophistication of that described above is, in all of our experience, habitually associated with intelligent agency. This brief description barely amounts the tip of the iceberg. Later, I will examine these intricate processes in more detail.
