 Evolution
Evolution
Parallel Host and Parasite Phylogenies: Do They Imply Common Ancestry?

One line of evidence for common ancestry, which lies among the stronger of the relevant arguments, is the phenomenon of parallel phylogenetic patterns thought to indicate host-parasite codivergence. Consider, for example, this diagram — or “tanglegram” as it’s called — excerpted from Legendre et al. (2001).
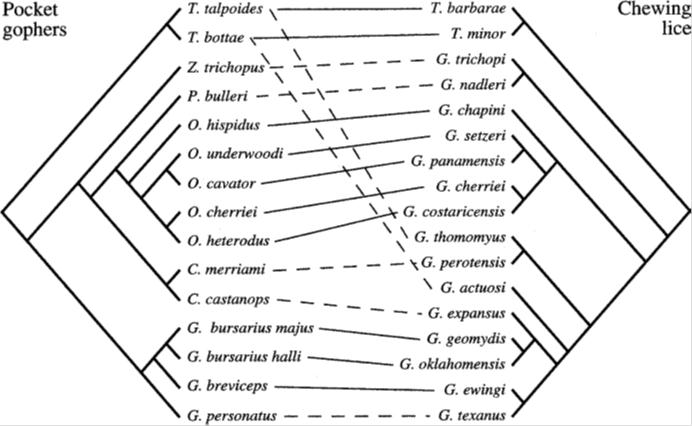
The tanglegram represents the parallel phylogenies yielded by pocket gophers (pictured above) and their lice parasites. The lines in the middle connect each species of pocket gopher to their corresponding parasites. As can be seen, with rare exceptions, the alignment of the trees is remarkably good. Occasionally, one observes a host-switching event. Similar studies have been conducted for quite a number of other groups, such as the host-parasite relationship between figs and fig wasps (R�nsted et al., 2005), or the relationship between parasites and birds (Ricklefs et al., 2004; Waldenstr�m et al., 2002), as well as primates (Garamszegi et al., 2009; Switzer et al., 2005) and also fish (Desdevises et al., 2002).
Sounds like a convincing argument for common descent, right? After all, the sheer number of potential phylogenies for those groups voids any appeal to chance. For this reason, it is widely thought that common ancestry and codivergence provide the only conceptual framework for understanding those parallel phylogenies.
Few would be surprised to learn that various species of pocket gophers are related. However, are there instances where such a pattern can be explained by some means other than shared ancestry of the host and its cospeciation with the parasite? Furthermore, are there cases where such a pattern defies explanation by common descent?
In 2000, Sharp et al. published a paper on “Origins and evolution of AIDS viruses: estimating the time-scale.” The team of researchers sought to determine the divergence times of primate lentivirus (a genus of retrovirus that includes HIV and SIV). Remarkably, they found that, while the lentivirus phylogeny closely paralleled the primate phylogeny, the divergence times were wildly different. The authors reported,
Some of the earliest attempts [to estimate the time-scale of primate lentivirus evolution], in 1988, yielded dramatically discordant results. One attempt to use a molecular clock estimated that the common ancestor of HIV-1 and HIV-2 existed as recently as 1951 [15]. At the same time, after comparison of HIV-1 with an SIV from an African green monkey, others suggested that these viruses may have diverged `gradually in concert with the evolution of primates’ [16], and that it was possible that a common ancestor of these viruses infected the common ancestor of apes and Old World monkeys [17]; that ancestor is thought to have lived at least 25 million years ago. Since HIV-1, HIV-2 and SIVAGM each belong to different major lineages of primate lentivirus (Figure 1), these two estimates pertain to the same common ancestor of all the primate lentiviruses. Thus the two different approaches led to estimates with a discrepancy of about six orders of magnitude! [emphasis added]
In the case outlined above, we are again faced with the sheer improbability of arriving at the parallel phylogenies by chance. Some other explanation is called for. Because the divergence times fail to correlate, we can also rule out cospeciation as an explanation.
How, then, might this discrepancy in fact be explained? Two main proposals have been put forward. One suggests that our modern molecular dating methodologies are faulty. While molecular dating is fraught with many problems, the other suggestion, in my opinion the more plausible, was offered in 2002 by Charleston and Robertson in a paper entitled “Preferential Host Switching by Primate Lentiviruses Can Account for Phylogenetic Similarity with the Primate Phylogeny.” They wrote,
If codivergence is insufficient to explain the similarity between the virus and host phylogenies, could any other process generate such coincidental results? We present here an alternative hypothesis of preferential host switching among primate host lineages, with host switching more likely to be successful between more closely related hosts than between more distantly related hosts. This hypothesis is used in a simulation study in which an artificial virus phylogeny is “grown” on the primate host phylogeny by a process of divergence (not codivergence) and host switching. We demonstrate how this simple principle can lead to artificially similar looking host and virus phylogenies, which can erroneously suggest codivergence where none occurred. [emphasis added]
It thus appears to be the case that cross-species transmission occurs preferentially between genetically similar species, thus producing the illusion of host-parasite cospeciation. The authors suggest that “There may well be other cases of host/parasite coevolution in which this phenomenon occurs.”
This phenomenon adds but one further example to a growing trend. While the scientific foundations of the modern evolutionary synthesis have been all but dashed to pieces by the relentless advance of science over the last decade or two, the scientific case for common descent is now too beginning to unravel.
Photo credit: David Hofmann.
