 Evolution
Evolution
 Faith & Science
Faith & Science
 Intelligent Design
Intelligent Design
 Life Sciences
Life Sciences
Michael Behe Hasn’t Been Refuted on the Flagellum
Those of us who have been reading the literature surrounding the ID/evolution controversy for any length of time will be quite acquainted with the standard Darwinian retort with regards the “Behean” argument for irreducible complexity as far as the bacterial flagellum is concerned. There seems to be this unanimity of opinion among Darwinian theorists that the claims of irreducible complexity with respect to the bacterial flagellum have been refuted, and that we ID proponents are constantly shifting the goal posts, burying our heads in the sand, and generally clutching at straws.
Indeed, one person on Facebook recently remarked,
My main gripe with the ID proponents is that they never seem to give up. How many times do you need to be told that something is wrong before you’ll admit it? How many times does ID need to be refuted in the peer reviewed media before you’ll give it up as a lost cause? The bacterial flagellum irreducible complexity story is completely and utterly dead. It’s wrong. Get over it.
I recently brought up the flagellum, as a documented instance of irreducible complexity, at a lunch bar Q&A session on the science/faith intersect, and received responses to much the same effect.
But is this claim actually true? Has this argument been refuted by critics? About a year ago, I read Why Intelligent Design Fails – A Scientific Critique of the New Creationism (edited by Matt Young and Taner Edis). Chapter 5 of that book was contributed by Ian Musgrave and is titled “Evolution of the Bacterial Flagellum.” Targeted as a response to Michael Behe and William Dembski, Musgrave attempts to dispel of the notion of irreducible complexity once and for all. Reading his chapter, I recall being deeply unimpressed. On page 82 of the book, Musgrave offers us the following argument:
Here is a possible scenario for the evolution of the eubacterial flagellum: a secretory system arose first, based around the SMC rod and pore-forming complex, which was the common ancestor of the type-III secretory system and the flagellar system. Association of an ion pump (which later became the motor protein) to this structure improved secretion. Even today, the motor proteins, part of a family of secretion-driving proteins, can freely dissociate and reassociate with the flagellar structure. The rod- and pore-forming complex may even have rotated at this stage, as it does in some gliding-motility systems. The protoflagellar filament arose next as part of the protein-secretion structure (compare the Pseudomonas pilus, the Salmonella filamentous appendages, and the E. coli filamentous structures). Gliding-twitching motility arose at this stage or later and was then refined into swimming motility. Regulation and switching can be added later, because there are modern eubacteria that lack these attributes but function well in their environment.(Shah and Sockett 1995). At every stage there is a benefit to the changes in the structure.
Indeed, Mark Pallen and Nick Matzke make a very similar argument in their 2006 Nature Reviews article (a paper which was raised by an audience member during the recent UK Behe tour). Ken Miller is also reputed for routinely making similar claims regarding the flagellum’s evolution from the Type III Secretion System based largely on considerations of protein sequence homologies.
So, do these points succeed in laying to rest that pesky business of intelligent design once and for all? Well, actually no; they don’t. In fact, I submit that the arguments of all of the aforementioned gentlemen fundamentally trivialize several important issues.
First and foremost, it trivializes the sheer complexity and sophistication of the flagellar system — both its assembly apparatus, and its state-of-the-art design motif. Actually, the process by which the bacterial flagellum is self-assembled within the cell is so sophisticated that I have long struggled to convey it in an accessible way to lay-persons. Its core concepts are notoriously difficult to grasp for those not accustomed to thinking about the system or for those encountering it for the first time. But, at the same time, the mechanistic basis of flagellar assembly is so breath-takingly elegant and mesmerizing that the sheer engineering brilliance of the flagellar motor — and, indeed, the magnitude of the challenge it brings to Darwinism — cannot be properly appreciated without at minimum a cursory knowledge of its underpinning operations. Let’s take a peek.
The Self-Assembly of the Flagellar Apparatus
 The synthesis of the bacterial flagellum requires the orchestrated expression of more than 60 gene products. Its biosynthesis within the cell is orchestrated by genes which are organised into a tightly ordered cascade in which expression of one gene at a given level requires the prior expression of another gene at a higher level. The paradigm, or model, organism for flagellar assembly is Salmonella, a bacterium of the family Enterobacteriaceae. My discussion thus pertains principally to Salmonella, unless otherwise indicated.
The synthesis of the bacterial flagellum requires the orchestrated expression of more than 60 gene products. Its biosynthesis within the cell is orchestrated by genes which are organised into a tightly ordered cascade in which expression of one gene at a given level requires the prior expression of another gene at a higher level. The paradigm, or model, organism for flagellar assembly is Salmonella, a bacterium of the family Enterobacteriaceae. My discussion thus pertains principally to Salmonella, unless otherwise indicated.
The flagellar system in Salmonella has three classes of promoters (promoters are akin to a kind of molecular toggle switch which can initiate gene expression when recognised by RNA polymerase and an associated specialised protein called a “sigma factor”). These three classes of promoters are uninspirationally dubbed “Class I,” “Class II,” and “Class III.” This sequential transcription is coupled to the process of flagellar assembly. Class I contains only two genes in one operon (called FlhD and FlhC). Class II consists of 35 genes across eight operons (including genes involved in the assembly of the hook-basal-body and other components of the flagellum, as well as the export apparatus and two regulatory genes called “FliA” and “FlgM”). Those genes which are involved in the synthesis of the filament are controlled by the Class III promoters.
The Class I promoter drives the expression of a master regulator (particular to the Enterobacteriaceae of which Salmonella is a member) called “FldH4C2” (don’t worry about remembering it!). This enteric master regulator then turns on the Class II promoters in association with a sigma factor, ?70 (remember, I said that a sigma factor is a type of protein which enables specific binding of RNA polymerase to gene promoters). The Class II promoters are then responsible for the gene expression of the hook-basal-body subunits and its regulators, including another sigma factor called ?28 (which is encoded by a gene called FliA) and its anti-sigma factor, FlgM (anti-sigma factors, as their name suggests, binds to sigma factors to inhibit their transcriptional activity). The sigma factor ?28 is required to activate the Class III promoters. But here we potentially run into a problem. It makes absolutely no sense to start expressing the flagellin monomers before completion of the Hook-Basal-Body construction. Thus, in order to inhibit the ?28, the anti-sigma factor (FlgM) alluded to above inhibits its activity and prohibits it from interacting with the RNA polymerase holoenzyme complex. When construction of the Hook-Basal-Body is completed, the anti-sigma factor FlgM is secreted through the flagellar structures which are produced by the expression of the Class II hook-basal-body genes. The Class III promoters (which are responsible for the expression of flagellin monomers, the chemotaxis system and the motorforce generators) are then finally activated by the ?28 and the flagellum can be completed.
But it gets better. The flagellar export system (that is, the means by which FlgM is removed from the cell) has two substrate-specificity states: rod-/hook-type substrates and filament-type substrates. During the process of flagellar assembly, this substrate-specificity switch has to flick from the former of those states to the latter. Proteins which form part of the hook and rod need to be exported before those which form the filament. But how does this switch in substrate-specificity take place?
A membrane-bound protein called FlhB is the key player in this process. There is also a flagellar hook-length protein which is responsible for making sure that the hook length is of the right size (around 55nm) called FliK. This same protein is also responsible for initiating the switch export substrate specificity. As it turns out, without FliK, both the ability to switch and export filament and the hook length control are completely lost. FliK has two key domains, i.e. the N-terminal and C-terminal domains. During hook assembly, FliKN functions as a molecular sensor and transmitter of information on hook length. When the hook reaches the right length, the information is transmitted to FliKC and FliKCT, resulting in a conformational change, which in turn results in FliKCT binding to FlhBC. This, in turn, results in a conformational change in FlhBC. This causes the substrate specificity switch.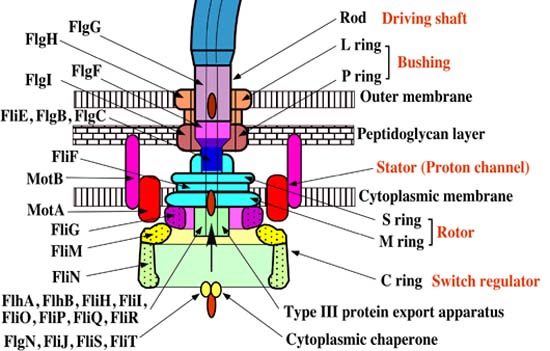
Flagellar assembly begins in the cytoplasmic membrane, progresses through the periplasmic space and finally extends outside the cell. The flagellum basically consists of two main parts: the secretion system and the axial structure. The principle components of the axial structure are FlgG for the rod, FlgE for the hook, and FliC for the filament. All of these assemble with the assistance of a cap protein (FlgJ, FlgD and FliD respectively). Of those, only FliD remains at the tip of the filament in the finished product. Other components of the axial structure (called FlgB, FlgC and FlgF) connect the rod and MS ring complex. The hook and filament are connected by FlgK and FlgL.
When the C ring and C rod attach to the M ring at its cytoplasmic surface, the MS ring complex — which is the structural foundation of the apparatus — can begin to secrete flagellar proteins.
The rod structure is built through the peptidoglycan layer. But its growth it isn’t able to proceed past the physical barrier presented by the outer membrane without assistance. So, the outer ring complex cuts a hole in the membrane, so that the hook can grow beneath the FlgD scaffold until it reaches the critical length of 55nm. Then the substrates which are being secreted can switch from the rod-hook mode to flagellin mode, FlgD can be replaced by hook-associated-proteins, and the filament continues to grow. Without the presence of the cap protein FliD, these flagellin monomers become lost. This cap protein is thus essential for the process to take place.
Why the Flagellum Evolution From the T3SS Doesn’t Work
One might have thought that the description given above should be more than enough to render the hand-waving gestures of Kenneth Miller et al. trivializations in the extreme. But it gets even worse for the Darwinian story. Why exactly is flagellum biosynthesis so tightly regulated and orchestrated? Not only do the energy demands render the flagellum an extremely expensive system to run, but untimely expression of flagellum proteins may induce a strong immune response in the host system, something no bacterium wants to do.
What is the significance of this from the standpoint of evolutionary rationale? Well, flagellin monomers are somewhat potent cytokine inducers. If you are a Yersinia organism in possession of a Type-III Secretion System the last thing you want to do is display those flagellin peptides to the macrophages. Such would no doubt be detrimental to the Yersinia’s anti-inflammatory mechanisms.
Conclusion
My description, given above, has really only scratched the surface of this spectacular item of nano-technology (for more detail, see here). I have not, for the sake of brevity, even discussed the remarkable processes of chemotaxis, two component signal transduction circuitry, rotational switching, and the proton motive force by which the flagellum is powered (for details on this, see my discussion here or, for more detail, see this review paper). But the bottom line is that modern Darwinian theory — as classically understood — has come no where close to explaining the origin of this remarkably complex and sophisticated motor engine. Just as Darwinian “explanations” of the eye may, at first, appear convincing to the uninitiated, largely unacquainted with the sheer engineering marvel of the biochemistry and molecular basis of vision, so too do the evolutionary “explanations” of the flagellum rapidly become void of any persuasiveness when one considers the molecular details of the system. When one couples the above details with demonstrations of the sheer impotence of neo-Darwinism to produce novel protein folds and novel protein-protein binding sites, do you really think that this system can be cobbled together by virtue of slight, successive modification, one small step at a time? Given that neo-Darwinism’s key selling point lies in its purported efficacy in explaining away the overwhelming appearance of design, doesn’t it stand to reason that its demonstrable impotence throws the design postulate back on the table as a viable and respectable scientific proposition?
Douglas Axe of the Biologic Institute showed in one recent paper in the journal Bio-complexity that the model of gene duplication and recruitment only works if very few changes are required to acquire novel selectable utility or neo-functionalization. If a duplicated gene is neutral (in terms of its cost to the organism), then the maximum number of mutations that a novel innovation in a bacterial population can require is up to six. If the duplicated gene has a slightly negative fitness cost, the maximum number drops to two or fewer (not inclusive of the duplication itself).
It seems that the bacterial flagellum is as much a — and perhaps a greater — challenge to Darwinism as it was when Behe first wrote Darwin’s Black Box in 1996.
