 Evolution
Evolution
 Life Sciences
Life Sciences
Testing the Orchard Model and the NCSE’s Claims of “Nested Patterns” Supporting a “Tree of Life”
In my previous post responding to the National Center for Science Education’s (NCSE) attacks on Explore Evolution‘s treatment of biogeography, I explained that there are many examples where there is inconsistency between evolutionary expectations of biogeography and plate tectonics. The NCSE is thus wrong to have claimed that “The consistency of these sorts of nested patterns cannot be explained without reference to common descent. The creationist ‘orchard’ is scientifically meaningless, since it makes no predictions.” * The classical “universal common descent” view is contrasted with the orchard model at below:
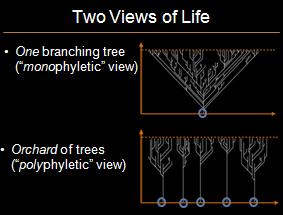
The NCSE’s claim is perplexing because, as noted, the NCSE also claimed that “continuity [between biogeographic and evolutionary patterns] is what would be expected of a pattern of common descent, and is not what would be expected with the creationist orchard scheme.” (emphasis added) Ignoring the NCSE’s inappropriate use of the “creationist” label, the NCSE seems to be suggesting that the orchard model is contradicted by the data. The NCSE is thus committing the classic evolutionist fallacy of arguing that opposing views are both unfalsifiable, and falsified by the data.
Regardless, the NCSE is wrong when it claims that the orchard model makes no predictions. If a monophyletic view of common descent predicts “nested patterns,” then by the NCSE’s own admission, a polyphyletic or “orchard” view predicts non-nested patterns. Indeed, systematists regularly search for precisely such non-nested patterns in order to identify polyphyletic taxa, a phenomenon effectively predicted by the orchard model. The only idea here that is “meaningless” is the NCSE’s claim that universal common descent makes predictions, while the “orchard” model does not (and, by the way, is falsified due to its failed predictions).
Biogeography is full of incongruent patterns which essentially entail non-nested distribution of species. In fact, Bruce S. Lieberman’s treatise, Paleobiogeography: Using Fossils to Study Global Change, Plate Tectonics, and Evolution, compares the problem of finding incongruent (i.e., non-nested) patterns among different biogeographic hypotheses to the problem of finding incongruent (i.e., non-nested) patterns of traits in different species when constructing phylogenetic trees:
[H]istorical biogeography is the discipline that looks at how groups of organisms have evolved and how their geographic distributions have changed in relation to geological or climatic events. … In phylogenetic analysis, the arbiter among competing hypotheses suggested by different character systems, i.e. incongruence among characters, is parsimony. The analogous problem in biogeography is what to do when one group suggests one biogeographic pattern, and another group suggests another.103
Analagous to maximizing parsimony in tree-construction, Lieberman notes that vicariance, or land-based separation of organisms, is the preferred explanation, such that the model that can “maximize vicariance, is the one preferred.”104 But Lieberman notes that there can be “incongruence”105 between biogeographic patterns. In Lieberman’s words, when “one group suggests one biogeographic pattern, and another group suggests another,” we have contradictory biogeographical data and we find the opposite of the NCSE’s claimed “continuity” that supports universal common descent.
In this regard, much of the data discussed in my previous post (and the “seamonkey post“) entails such incongruence and a breakdown in nested patterns of biogeographic distribution of taxa. As seen, such disparate data often require evolutionists to resort to speculative and unfalsifiable hypotheses of oceanic dispersal as a means of transcending traditional methods of migration. This data challenges the “continuity” of biogeographic and evolutionary patterns said to support universal common descent, but it might be expected under an orchard model.
In fact, it is not only within biogeography that we find non-nested patterns, and it important to fact-check the NCSE’s claim that we always find “nested patterns” pointing to a “tree of life.” There’s a January 2009 article in New Scientist titled, “Why Darwin Was Wrong about the Tree of Life.” Contrary to the NCSE’s claim that we always find “nested patterns” which “cannot be explained without reference to common descent,” the article reported a major “problem” encountered by molecular systematists, namely that “different genes told contradictory evolutionary stories.” The article observed that with the sequencing of the genes and proteins of various living organisms, the tree of life fell apart:
“For a long time the holy grail was to build a tree of life,” says Eric Bapteste, an evolutionary biologist at the Pierre and Marie Curie University in Paris, France. A few years ago it looked as though the grail was within reach. But today the project lies in tatters, torn to pieces by an onslaught of negative evidence. Many biologists now argue that the tree concept is obsolete and needs to be discarded. “We have no evidence at all that the tree of life is a reality,” says Bapteste. That bombshell has even persuaded some that our fundamental view of biology needs to change.106
Of course, these scientists are all committed evolutionists, which makes their admissions all the more weighty. And these arguments apply not just to the base of the tree of life, but also to higher branches where processes like horizontal gene transfer are not thought to be prevalent, as the article observed that “the evolution of animals and plants isn’t exactly tree-like either.”
To reiterate, the basic problem is that one gene or protein yields one version of the “tree of life,” while another gene or protein yields an entirely different tree. As the New Scientist article stated:
The problems began in the early 1990s when it became possible to sequence actual bacterial and archaeal genes rather than just RNA. Everybody expected these DNA sequences to confirm the RNA tree, and sometimes they did but, crucially, sometimes they did not. RNA, for example, might suggest that species A was more closely related to species B than species C, but a tree made from DNA would suggest the reverse.107
Likewise, leading evolutionary bioinformatics specialist W. Ford Doolittle explains, “Molecular phylogenists will have failed to find the ‘true tree,’ not because their methods are inadequate or because they have chosen the wrong genes, but because the history of life cannot properly be represented as a tree.”108 The NCSE may claim that this problem is only encountered when one tries to reconstruct the evolutionary relationships of microorganisms, such as bacteria, which can swap genes through a process called “horizontal gene transfer,” thereby muddying any phylogenetic signal. But the tree of life is challenged even among higher organisms where such promiscuous gene-swapping across taxa is not thought to not take place. As the article explains:
Syvanen recently compared 2,000 genes that are common to humans, frogs, sea squirts, sea urchins, fruit flies and nematodes. In theory, he should have been able to use the gene sequences to construct an evolutionary tree showing the relationships between the six animals. He failed. The problem was that different genes told contradictory evolutionary stories. This was especially true of sea-squirt genes. Conventionally, sea squirts–also known as tunicates–are lumped together with frogs, humans and other vertebrates in the phylum Chordata, but the genes were sending mixed signals. Some genes did indeed cluster within the chordates, but others indicated that tunicates should be placed with sea urchins, which aren’t chordates. “Roughly 50 per cent of its genes have one evolutionary history and 50 per cent another,” Syvanen says.109
Even among higher organisms, “[t]he problem was that different genes told contradictory evolutionary stories,” leading Syvanen to say, regarding the relationships of these higher groups, “We’ve just annihilated the tree of life.” This directly contradicts the NCSE’s claim that there exists a “tree of life” with “nested patterns” which “cannot be explained without reference to common descent.”
Other scientists agree with the conclusions of the New Scientist article. While many try to explain phylogenetic conflicts as being the result of gene swapping among microorganisms at the base of the tree of life, problems with the tree of life extend all the way up the tree. Likewise, Carl Woese, a pioneer of evolutionary molecular systematics, observed that these problems extend well beyond the base of the tree of life: “Phylogenetic incongruities [conflicts] can be seen everywhere in the universal tree, from its root to the major branchings within and among the various taxa to the makeup of the primary groupings themselves.”111
National Academy of Sciences biologist Lynn Margulis has had harsh words for the field of molecular systematics, which Hillis studies. In her article, “The Phylogenetic Tree Topples,” she explains that “many biologists claim they know for sure that random mutation (purposeless chance) is the source of inherited variation that generates new species of life and that life evolved in a single-common-trunk, dichotomously branching-phylogenetic-tree pattern!” But she dissents from that view and attacks the evolutionary systematists, noting, “Especially dogmatic are those molecular modelers of the ‘tree of life’ who, ignorant of alternative topologies (such as webs), don’t study ancestors.”112
Striking admissions of troubles in reconstructing the “tree of life” also came from a paper in the journal PLoS Biology entitled, “Bushes in the Tree of Life.” The authors acknowledge that “a large fraction of single genes produce phylogenies of poor quality,” observing that one study “omitted 35% of single genes from their data matrix, because those genes produced phylogenies at odds with conventional wisdom.”113 The paper suggests that “certain critical parts of the [tree of life] may be difficult to resolve, regardless of the quantity of conventional data available.”114 The paper even contends that “[t]he recurring discovery of persistently unresolved clades (bushes) should force a re-evaluation of several widely held assumptions of molecular systematics.”115
Unfortunately, one assumption that these evolutionary biologists are not willing to consider changing is the assumption that neo-Darwinism and universal common ancestry are correct. Meanwhile, as far as the data are concerned, the New Scientist article admits, “Ever since Darwin the tree has been the unifying principle for understanding the history of life on Earth,” but because “different genes told contradictory evolutionary stories,” the notion of a tree of life is now quickly becoming a vision of the past — as the article stated, it’s being “annihilated.”
The NCSE claims that the “orchard” concept of EE is “meaningless” but it would seem to predict the precise non-nested phylogenetic data reported in New Scientist and the non-nested biogeographic data reported in Part III above. Perhaps the reason why different genes are telling “different evolutionary stories” and “one group suggests one biogeographic pattern, and another group suggests another” is because the genes and organisms have wholly different stories to tell, namely stories that indicate that not all living organisms are ancestrally related, thereby fulfilling a testable prediction of the orchard model.
References Cited:
* All quotes of the NCSE in this document were downloaded from the NCSE website’s response to EE on Biogeography on October 29, 2008.
[103.] Bruce S. Lieberman, Paleobiogeography: Using Fossils to Study Global Change, Plate Tectonics, and Evolution, pg. 114 (Kluwer Academic Press, 2000) (emphasis added).
[104.] Id. at 124.
[105.] Id. at 135.
[106.] Graham Lawton, “Why Darwin was wrong about the tree of life,” New Scientist (January 21, 2009) (emphasis added).
[107.] Id.
[108.] W. Ford Doolittle, “Phylogenetic Classification and the Universal Tree,” Science, Vol. 284:2124-2128 (June 25, 1999).
[109.] Graham Lawton, “Why Darwin was wrong about the tree of life,” New Scientist (January 21, 2009).
[111.] Carl Woese “The Universal Ancestor,” Proceedings of the National Academy of Sciences USA, Vol. 95:6854-9859 (June, 1998) (emphasis added).
[112.] Lynn Margulis, “The Phylogenetic Tree Topples,” American Scientist, Vol 94 (3) (May-June, 2006).
[113.] Antonis Rokas & Sean B. Carroll, “Bushes in the Tree of Life,” PLoS Biology, Vol 4(11): 1899-1904 (Nov., 2006) (internal citations and figures omitted).
[114.] Id.
[115.] Id.
